NEEDLEMAN-WUNSCH AND SMITH-WATERMAN COMBINATIONS IN PAIRWISE ALIGNMENT
Abstract
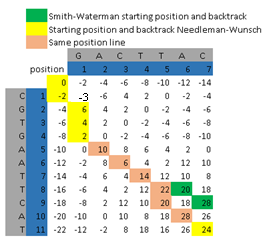
Identification of Deoxyribonucleic acid (DNA), Ribonucleic acid (RNA), or protein needs to be done to find functional, structural, or evolutionary relationships between two sequences. There are various applications that already exist such as one of them EMBOSS either web or desktop versions. There are drawbacks to this application, such as repeatedly processing each user who needs sequence alignment results locally and globally at the same time. Therefore, we designed an application that can generate two sequence alignment outputs both locally and globally at the same time with pairwise alignment Needleman-Wunsch and Smith-Waterman. The result show that methods can be produces two outputs of sequence alignment in the same process. The impact is it can reduce the waiting time for users.
Downloads
Copyright (c) 2024 Jurnal Mnemonic

This work is licensed under a Creative Commons Attribution-ShareAlike 4.0 International License.

















